Main research topic: cell fragmentation
What is cell fragmentation?
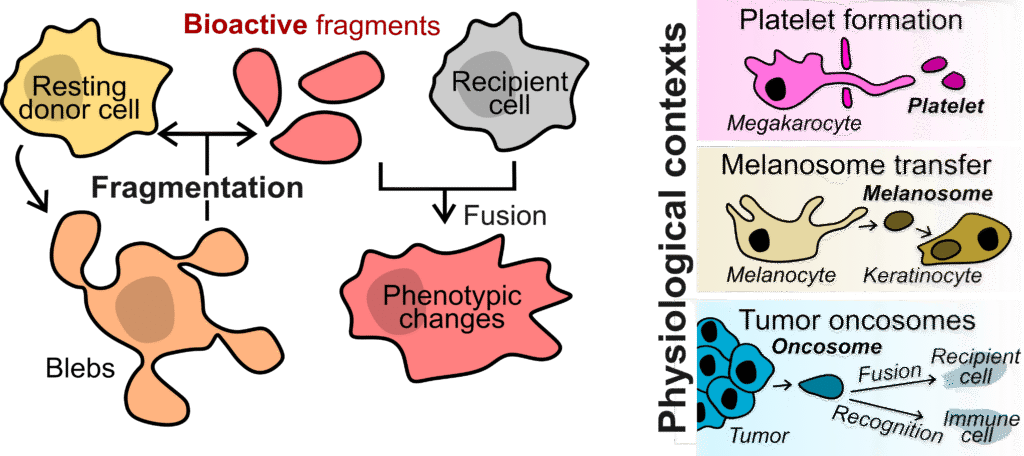
Cell fragmentation is the partition of a cell into two entities outside the context of cell division. Many cell types partition physiologically, producing large bioactive fragments without a nucleus that make up for 5-30% of the initial cell volume. These fragments can retain biological activity for several days without transcription (e.g., platelets from megakaryocytes), fuse with a recipient cell and thereby transfer organelles (e.g., melanosomes transferred from melanocytes to keratinocytes) or activate signaling and metabolic pathways in the recipient cell (e.g., oncosomes from cancer cells). Despite profound implications in health and disease, cell fragmentation is currently poorly understood from a quantitative and mechanistic perspective.
Significance
Cells can shed large pieces of themselves that remain functional and influence their surroundings. This process, called cell fragmentation, appears in many settings but is still poorly understood. By uncovering the physical and molecular rules that generate these cytoplasts, we aim to clarify how cells respond to mechanical stress, communicate within tissues, and shape processes such as cancer progression and platelet formation. Understanding fragmentation will give us a broader view of how cells share, reorganize, and repurpose their cytoplasm in health and disease.
Research lines
1. Physical principles of fragmentation
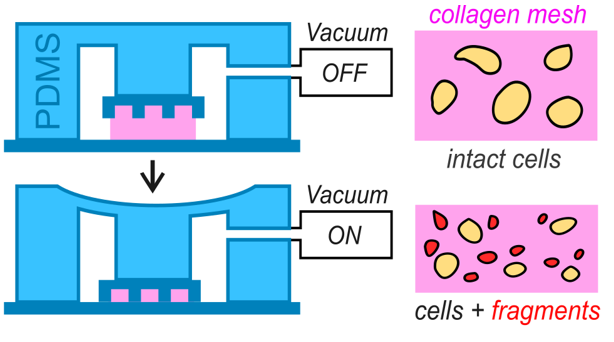
We study how forces, confinement, membrane tension, and cortical dynamics produce thin bleb necks that can constrict and sever. Using microfluidic confinement systems, we define the mechanical conditions that favor fragmentation and identify the minimal ingredients needed for cytoplast release, in collaboration with theoreticians (Voituriez group, UPMC Paris).
2. Reconstituting fragmentation in vitro
We use our microfluidic devices to reconstitute fragmentation under controlled geometry and biochemical cues. This allows us to isolate core mechanisms and quantify contractility, membrane behavior, and scission events with quantitative live imaging.
3. Composition and function of fragments
Fragments carry selected proteins, lipids, metabolites, and sometimes organelles. We map this content using quantitative imaging, metabolomics, and lipidomics (with the neighbor D’Angelo lab at EPFL) to understand what fragments retain and how this shapes their stability and behavior.
4. Fragmentation in cancer
Cancer cells in confined tissues produce large cytoplasts that migrate, signal, and interact with immune cells. We explore how tumor mechanics promote fragmentation and how these cytoplasts influence cancer progression and immune responses, working closely with in vivo collaborators (Friedl group at Radboud University Netherlands, and clinicians from the local hospital CHUV by the TUMORSHARE platform).
Other topics we are interested in…
Microfluidics design
We design microfluidic systems to impose controlled mechanical and chemical landscapes on cells. These devices allow us to reconstitute specific physical constraints, shape cell behaviour, and generate fragmentation events in vitro. Our approach combines precision engineering with quantitative imaging to create experimental platforms that reveal how cells respond to defined forces and geometries.
Link to review: ‘Reconstitution of cell migration’
Link to methods: ‘Extended methods for 2D confinement’
Cell volume regulation
Cells constantly adapt their volume in response to mechanical stress and osmotic fluctuations. We are interested in how water flux, ion transport, and membrane tension interact with the actin cortex to produce dynamic volume changes. Our interest includes also oscillatory osmotic shocks, coupling between membranes and the cytoskeleton, and how volume regulation influences migration, fragmentation, and cell state transitions.
Fluorescence Lifetime Imaging Microscopy
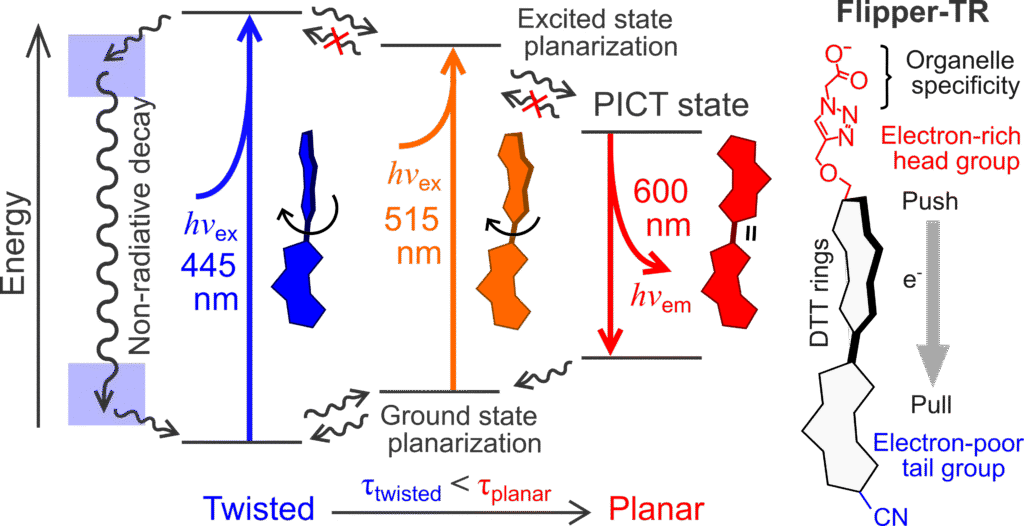
Our lab has contributed to the development of technical standards and quantitative methods for FLIM. We are interested in applying FLIM-based sensors such as Flipper-TR to measure physical and molecular properties at sub-cellular level, including membrane tension, molecular crowding, and local changes in lipid organization.
Link to method paper: ‘Fluorescence Lifetime Estimation’
Blebology and cell migration
We are interested in how non-apoptotic blebs form, grow, and resolve, and how these membrane protrusions lead to cell migration. Blebs provide a simple interface to study membrane tension, cortex dynamics, and actomyosin regulation. Our work on my migration focuses on the role of membrane tension, cortical dynamics, and substrate interaction during migration in confinement. We are interested in how these physical parameters shape directional cell migration.